- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

HighLights

Arul Chinnaiyan
American Cancer Society Research
Professor
DIRECTOR, Michigan center for
Translational Pathology
Investigator, Howard Hughes Medical
Institute
Professor of Pathology and Urology
S.P. Hicks Endowed Professor
of Pathology
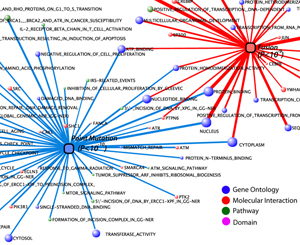 INTEGRATIVE ANALYSIS OF
INTEGRATIVE ANALYSIS OF
‘-OMICS’ IN CANCER
Despite great advances with microarrays, the rapid evolution of Next Generation Sequencing (NGS) has enabled an unbiased view of a cancer’s genome, transcriptome, and methylome, as in The Cancer Genome Atlas (TCGA) program. However, the impact of high-throughput genomics technologies on diagnosis, prognosis, and novel therapies will ultimately be determined by the ability to integrate, visualize, and interpret the streams of data. Therefore, we plan to establish a platform for integrative ‘-omics’ analysis to comprehensively catalog alterations occurring throughout tumor progression. We believe an integrative approach will enable us to distinguish causative events, hidden amongst private non-specific alterations that would serve as the basis for novel targeted therapies, especially in the most aggressive metastatic cancers, that will ultimately lead to improved patient survival.
Overall Goals
- Utilize Next Generation sequencing to elucidate systems level modifications and their subsequent effect on regulatory networks and pathways t.o promote more aggressive forms of cancer.
- To elucidate transcriptional aberrations that can serve as valuable therapeutic targets..
Themes and Specific Aims
- Comprehensively catalog genomic, epigenomic, and transcriptomic alterations during cancer progression based on next-generation sequencing. We will leverage existing analysis methods to build pipeline workflows for individual NGS technologies that facilitate the downstream integration described below.
- Integrate multi-dimensional results from NGS experiments and from other molecular levels to identify mechanisms driving tumor progression. We will characterize the levels and changes in DNA, mRNA, proteins, micro-RNA, epigenome, and metabolites in tumor tissue and cancer cell lines to elucidate mechanisms driving tumor differentiation in metastatic cancers. We expect to identify and characterize pathways and regulatory modules that can inform choices of potential biomarker candidates and targets for therapy.
Related Publications
Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature, 2009: 458(7234): 97-101.
Maher, CA., Palanisamy, N., Brenner, J.C., Cao, X., Kalyana-Sundaram, S., Luo, S., Khrebtukova, I., Barrette, T., Quist, C., Yu, J., Lonigro, R., Schroth, G., Kumar-Sinha, C., and Chinnaiyan, A. (2009) Chimera transcript discovery using paired-end transcriptome sequencing. PNAS 106(30):12353-8.
Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008; 68(3): 645-9. PMID: 18245462.
Taylor BS, Pal M, Yu J, Laxman B, Kalyana-Sundaram S, Zhao R, Menon A, Wei JT, Nesvizhskii AI, Ghosh D, Omenn GS, Lubman DM, Chinnaiyan AM, Sreekumar A. Humoral response profiling reveals pathways to prostate cancer progression. Mol Cell Proteomics 2008; 7(3): 600-11. PMID: 18077443.
Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, Demichelis F, Helgeson BE, Laxman B, Morris DS, Cao Q, Cao X, Andrén O, Fall K, Johnson L, Wei JT, Shah RB, Al-Ahmadie H, Eastham JA, Eggener SE, Fine SW, Hotakainen K, Stenman UH, Tsodikov A, Gerald WL, Lilja H, Reuter VE, Kantoff PW, Scardino PT, Rubin MA, Bjartell AS, Chinnaiyan AM. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 2008; 13(6): 519-28. PMID: 18538735.
Morris DS, Tomlins SA, Rhodes DR, Mehra R, Shah RB, Chinnaiyan AM. Integrating biomedical knowledge to model pathways of prostate cancer progression. Cell Cycle 2007; 6(10): 1177-87. PMID: 17495538.
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007; 9(2): 166-80. PMID: 17356713.
Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007; 39(1): 41-51. PMID: 17173048.
Mo F, Hong X, Gao F, Du L, Wang J, Omenn GS, Lin B. A compatible exon-exon junction database for the identification of exon skipping events using tandem mass spectrum data. BMC Bioinformatics (in press).
Tu LC, Yan X, Hood L, Lin B. Proteomics analysis of the interactome of N-myc downstream regulated gene 1 and its interactions with the androgen response program in prostate cancer cells. Mol Cell Proteomics 2007; 6(4): 575-88. PMID: 17220478.
Lin B, White JT, Lu W, Xie T, Utleg AG, Yan X, Yi EC, Shannon P, Khrebtukova I, Lange PH, Goodlett DR, Zhou D, Vasicek TJ, Hood L. Evidence for the presence of disease-perturbed networks in prostate cancer cells by genomic and proteomic analyses: a systems approach to disease. Cancer Res 2005; 65(8): 3081-91. PMID: 15833837.
Yu J, Yu J, Almal AA, Dhanasekaran SM, Ghosh D, Worzel WP, Chinnaiyan AM. Feature selection and molecular classification of cancer using genetic programming. Neoplasia 2007; 9(4): 292-303. PMID: 17460773.
Tomlins SA, Chinnaiyan AM. Of mice and men: cancer gene discovery using comparative oncogenomics. Cancer Cell 2006; 10(1): 2-4. PMID: 16843259.
Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res 2006; 66(7): 3396-400. PMID: 16585160.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310(5748): 644-8. PMID: 16254181.
Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiyan AM. Autoantibody signatures in prostate cancer. N Engl J Med 2005; 353(12): 1224-35. PMID: 16177248.

 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

