- Search NCIBI Data
(e.g. diabetes, csf1r) - Login

 Computational Advances
Computational Advances
Computational systems biology and medicine require fresh approaches to manage and intelligently process the ever increasing amount of biomedical data generated by technological advances. In computer science, NCIBI specialists are addressing the computational, database, data mining, visualization, data sharing, and mathematical modeling needs of the Center’s Driving Biological Projects. The goal is to facilitate and enhance analyses conducted by clinical and translational researchers - important end-users of these technologies. Key technical contributions include the following:
Tool Integration
We aim for interoperable suite of tools and data resources for multiscale systems analysis through architectures that target scientists’ needs for a uniform search interface across all NCIBI tools and data resources.
- Workspaces that provide richer contexts than just the last gene visited.
- A “list of X” architecture for genes, proteins, metabolites, articles, concepts, and network topology underlies the workplaces. With it, scientists, for example, can examine lists of differentially expressed genes and group them by relevant sets of enriched concepts derived from the corpus of Medline literature.
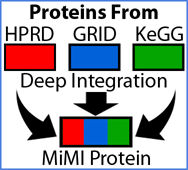
Deep Data Integration
Effective biomedical research requires convenient access to information that is multi-scaled, from many sources, and in diverse modalities. Towards these ends, we:
- Collect and organize relevant data from numerous sources from each molecular and physiologic level and stage.
- Filter actionable data into information
- Prepare the data and information for mining. Scientists’ queries return relationships and patterns to accelerate hypothesis testing and knowledge creation.
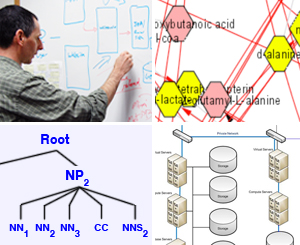
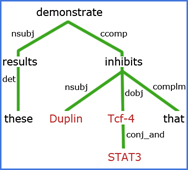
Natural Language Processing (NLP)
In applying NLP, challenges that we address in extracting facts and related context from biomedical literature include:
- Named entity extraction, entity disambiguation, mined ontology extraction, anaphora resolution, and speculative statements.
- Tagging genes, metabolites, and other biomedically important concepts in literature, allowing scientists to make inferences of new (currently unknown) information through systematic analysis, for example, between lists of MeSH terms and lists of genes/metabolites.
Visualization
NCIBI has worked closely with Cytoscape to extend features of this popular tool and develop a Cytoscape plug-in for MiMI and MetScape. Cytoscape and our plug-ins facilitate dynamically exploring and analyzing high dimensional molecular interaction networks.
Examples of successes include:
- Cytoscape Plug-in for MiMI
- The NCIBI Cytoscape plugin for MiMI queries the MiMI molecular interactions database, incorporates the NLP tools SAGA and LexRank, and integrates with MiMI Web interface and other NCIBI Tools (Gao et al., 2009).
- Cytoscape plug in for Metabolomics: Metscape
- The NCIBI Metscape plug-in module visualizes compound interactions and reactions; places them in the context of metabolic networks; and animates scientists’ time series experimental data to reveal changes in user-specified values (Gao et al., submitted).
- Web-based Tools for Accessing our BioNLP data
- NCIBI has many tools that use our fully relational, up-to-date biomedical text database drawing data from PubMed, PubMedCentral and full text sources. Tools which utilize these data include MiSearch, Gene2MeSH, Metab2MeSH, GIN, and the NLP web service.
- ConceptGen
- ConceptGen is a tool that finds relationships between biomedical concepts based on the genes they have in common. For example, given a user-defined set of genes of interest (e.g. from microarray results), ConceptGen suggests biological concepts that may be relevant to this set, and hence serve as potential loci for further exploration.


 NCIBI on Facebook
NCIBI on Facebook NCIBI RSS Feed
NCIBI RSS Feed

